Explainable artificial intelligence (xAI) meets drug discovery
Various concepts of artificial intelligence (AI) have been successfully applied to computationally assisted drug discovery over the past few years.
This advancement is largely due to deep learning algorithms, artificial neural networks with multiple processing layers that are able to model complex nonlinear input-output relationships and perform pattern recognition and feature extraction from low-level data representations.
Certain deep learning models have been shown to match or exceed the performance of existing machine learning and quantitative structure-activity relationship (QSAR) methods in drug discovery.
In addition, deep learning also enhances the potential of computer-aided discovery and broadens its scope of application, such as molecular design, chemical synthesis design, protein structure prediction, and macromolecular target identification.
The ability to capture complex nonlinear relationships between input data and associated outputs often comes at the expense of limited comprehensibility of the resulting model. Despite ongoing efforts in algorithmic interpretation and analysis of molecular descriptors to explain QSAR, deep neural network models notoriously evade direct human access. Therefore, blurring the line between the "two QSARs" (ie, machine-learning interpretable models and highly accurate models) may be the key to accelerating drug discovery with AI.
Automated analysis of medical and chemical knowledge to extract and represent features in a human-understandable format dates back to the 1990s, but has received increasing attention due to the re-emergence of neural networks in chemistry and healthcare.
Given the current pace of AI development in drug discovery and related fields, there will be a growing need for methods to help researchers understand and interpret the underlying models.
To alleviate the lack of interpretability of some machine learning models and to enhance human reasoning and decision-making, explainable artificial intelligence (xAI) approaches have received attention.
While providing mathematical models, the purpose of providing informative explanations is to: make the underlying decision-making process transparent ("understandable"); avoid making correct predictions for the wrong reasons (the so-called smart Hans effect).
avoid unfair bias or unethical discrimination; bridge the gap between the machine learning community and other scientific disciplines. In addition, effective xAI can help scientists cross the "cognitive valley", allowing them to Hone your knowledge and beliefs in the research process.
Transparency, knowing how the system came to a particular answer.
Justification, stating why the answer provided by the model is acceptable.
Informative, providing new information to human decision makers.
Uncertainty, estimates the reliability of quantitative forecasts.
In general, the explanations produced by xAI can be classified as global or local. Furthermore, xAI can be dependent on the underlying model or be agnostic, which in turn affects the potential applicability of each approach. In this framework, there is no one-size-fits-all approach to xAI.
There are a number of domain-specific challenges for future AI-assisted drug discovery, such as the representation of data fed back to the method.
In contrast to many other fields in which deep learning has been shown to excel, such as natural language processing and image recognition, there is no naturally applicable, complete, "raw" molecular representation. After all, molecules are models themselves, as scientists envision them.
Therefore, this 'inductive' approach to building higher-order models from lower-order models is philosophically questionable.
The choice of molecular "characterization model" becomes a limiting factor for the interpretability and performance of the resulting AI model, as it determines the content, type, and interpretability of the retained chemical information.
Drug design is not simple. It differs from explicit engineering due to errors, non-linearities, and seemingly random events.
The incomplete understanding of molecular pathology and the inability to develop airtight mathematical models and corresponding explanations for drug action must be acknowledged.
In this context, xAI bears the potential to augment human intuition and skills to design novel bioactive compounds with desirable properties.
The design of new drugs is a matter of whether pharmacological activity can be deduced from the molecular structure, and which elements of this structure are relevant. Multi-objective design presents more challenges and sometimes unsolvable problems, resulting in molecular structures that are often compromises.
Practical approaches aim to limit the number of syntheses and assays required to find and optimize new hits and leads, especially when elaborate and expensive tests are performed. xAI-assisted drug design promises to help overcome some of these issues by taking informed action that takes into account medicinal chemistry knowledge, model logic, and awareness of system limitations.
xAI will facilitate collaboration between medicinal chemists, cheminformatics and data scientists.
In fact, xAI can already realize the mechanism explanation of drug action, and contribute to the improvement of drug safety, as well as the design of organic synthesis.
If successful in the long term, xAI will provide fundamental support for analyzing and interpreting increasingly complex chemical data, as well as for developing new pharmacological hypotheses, while avoiding human bias.
Pressured drug discovery challenges, such as the coronavirus pandemic, may spur the development of applied tailored xAI approaches to rapidly address specific scientific questions relevant to human biology and pathophysiology.
The field of xAI is still in its infancy, but is advancing at a rapid pace, and its relevance is expected to increase over the next few years.
In this review, the researchers aim to provide a comprehensive overview of recent xAI research, highlighting its strengths, limitations, and future opportunities for drug discovery.
In what follows, existing and some potential drug discovery applications are presented after a structured conceptual taxonomy of the most relevant xAI approaches.
Finally, the limitations of contemporary xAI are discussed and potential methodological improvements needed to facilitate the practical application of these techniques in pharmaceutical research are pointed out.
xAI:Technology status and future directions:
This section aims to provide a succinct overview of modern xAI methods, with examples of their applications in computer vision, natural language processing, and discrete mathematics.
Then, selected case studies in drug discovery will be highlighted, and potential future areas and research directions for xAI in drug discovery will be proposed.
In the following, without loss of generality, f will denote a model; x ∈ X will be used to denote the set of features describing a given instance that are used by f to make predictions y ∈ Y.
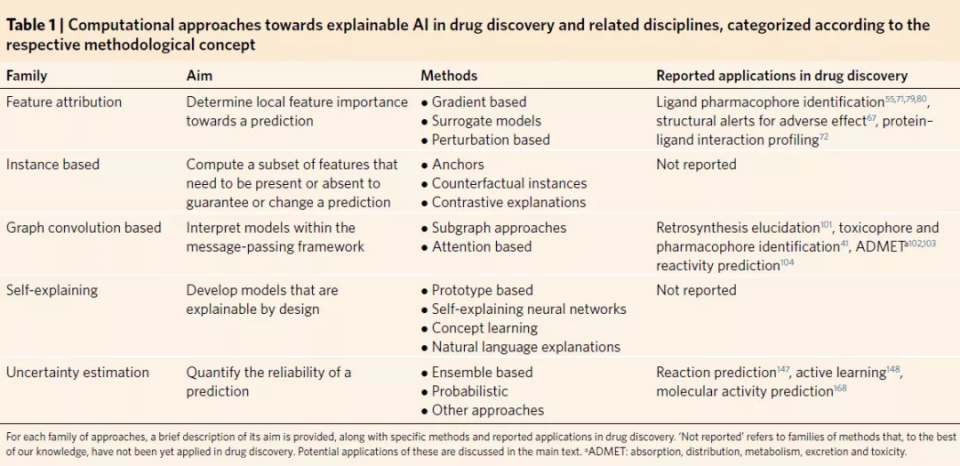
xAI:Feature Attribution Methods
Given a regression or classification model F: X ∈ [Rk → R (where [R] refers to the set of real numbers and K (as [R) refers to the k-dimensional set of real numbers), the feature attribution method is a function E: X ∈ [ Rk→[Rk It takes model input and produces an output whose value represents the relevance of each input feature to the final prediction computed with f.
Feature attribution methods can be divided into the following three categories.
Gradient-based feature attribution.
Proxy model feature attribution.
Perturbation-based methods aim to modify or remove parts of the input to measure its corresponding change in the model output; this information is then used to assess feature importance.
Over the past few years, feature attribution methods have been the most commonly used for ligand- and structure-based drug discovery in the xAI family of techniques.
It should be noted that the interpretability of feature attribution methods is limited by the original feature set.
Especially in drug discovery, the use of complex or "opaque" input molecular descriptors often hinders interpretability. When using feature attribution methods, it is recommended to choose understandable molecular descriptors or model building models.
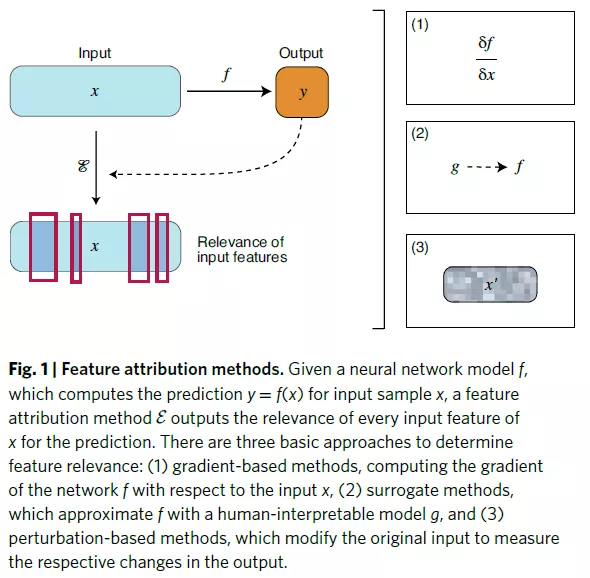
xAI for cytochrome P450-mediated metabolism
This working example demonstrates that xAI provides a graphical interpretation, in terms of molecular motifs, that are considered relevant by neural network models for predicting drug interactions with cytochrome P450s.
Comprehensive gradient feature attribution combined with graph convolutional neural networks for predicting drug interactions with CYP3A4.
The network model was trained with a set of published CYP3A4 substrates and inhibitors.
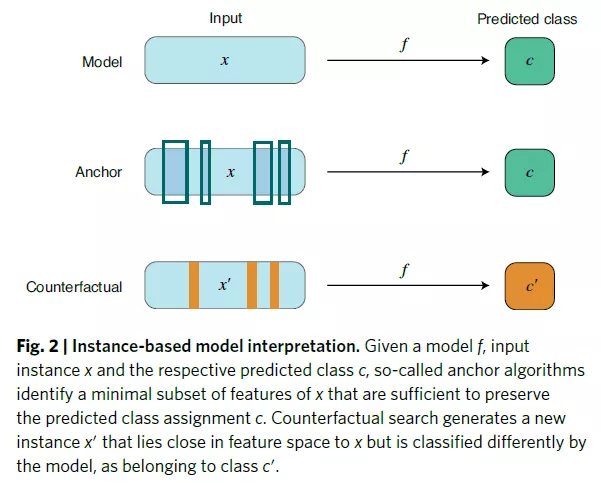
xAI:instance-based approach
Instance-based methods compute a subset of relevant features that must exist to preserve the predictions of a given model.
Instances can be real or generated for the purpose of the method. It has been argued that instance-based approaches provide humans with "natural" interpretations of models because they resemble counterfactual reasoning.
The anchor algorithm provides an interpretable interpretation of the classifier model.
Counterfactual instance search.
Contrastive interpretation methods provide instance-based interpretability of classifiers by generating sets of "relevant positives" and "relevant negatives".
In drug discovery, instance-based approaches may be valuable for increasing model transparency by highlighting the presence or absence of molecular features that need to warrant or alter model predictions.
Furthermore, counterfactual reasoning further increases the amount of information by exposing potentially new information about the model and underlying training data to human decision makers.
To the best of the authors' knowledge, instance-based approaches have not yet been applied to drug discovery. The authors believe they hold promise in several areas of de novo molecular design.
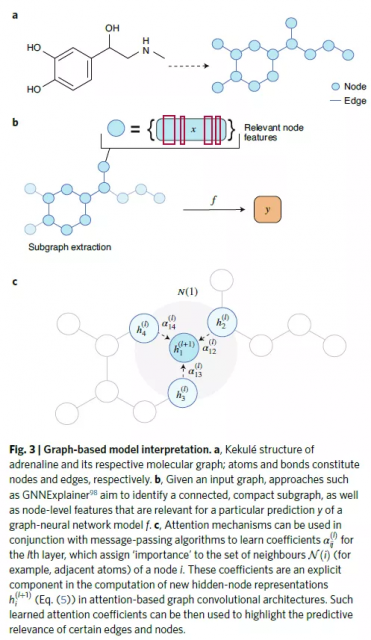
xAI:Methods based on graph convolution
Molecular graphs are natural mathematical representations of molecular topology, with nodes and edges representing atoms and chemical bonds, respectively.
Their use in cheminformatics and mathematical chemistry has been common since the late 1970s.
Therefore, it is not surprising to witness the increasing use of novel graph convolutional neural networks in these fields, which formally fall under the category of neural message passing algorithms.
Generally speaking, convolution refers to the mathematical operation of two functions to produce a third function that expresses how the shape of one is modified by the other.
This concept is widely used in convolutional neural networks for image analysis.
Graph convolution naturally extends convolution operations typically used in computer vision or natural language processing applications to graphs of arbitrary size.
In the context of drug discovery, graph convolutions have been applied in generative models for molecular property prediction and new drug design.
Exploring the interpretability of models trained with graph convolutional architectures is currently a particularly active research topic.
The graph convolution-based xAI methods in this paper fall into the following two categories.
Subgraph identification methods aim to identify one or more parts of the graph that are responsible for a given prediction.
Attention-based methods.
Interpretation of graph convolutional neural networks can benefit from attention mechanisms, taken from the field of natural language processing, where their use has become standard.
Graph convolution-based methods represent a powerful tool in drug discovery due to their direct and natural connection to chemists' intuitive representations.
Furthermore, when used in conjunction with knowledge, the possibility to highlight atoms relevant to specific predictions can improve model demonstration and its informative flow to fundamental biology and chemistry.
Due to its intuitive connection to two-dimensional representations of molecules, graph convolution-based xAI has the potential to be applied to several other common modeling tasks in drug discovery.
In the authors' opinion, xAI for graph convolution may be most beneficial for applications aimed at finding relevant molecular motifs, such as structural alert identification and sites for reactivity or metabolic prediction.
xAI:self-explaining method
The xAI methods introduced so far produce posterior interpretations of deep learning models.
While this post-hoc explanation has proven useful, some argue that, ideally, xAI methods should automatically provide human-interpretable explanations along with their predictions. Such an approach would facilitate validation and error analysis and could be directly linked to domain knowledge.
While the term self-explaining was coined to refer to a specific neural network architecture, in this review, the term is used in a broad sense to refer to approaches with interpretability at the core of their design.
Self-explanatory xAI methods can be divided into the following categories.
Prototype-based reasoning refers to the task of predicting future events based on particularly useful known data points.
Self-explanatory neural networks aim to associate input or latent features with semantic concepts.
Human-interpretable concept learning refers to the task of learning a class of concepts from data, aiming to achieve human-like generalization abilities.
A test with a concept activation vector computes the directional derivative of the layer's activation with respect to its input towards the concept direction. Such a derivative quantifies the degree to which the latter is relevant to a particular classification.
Natural language interpretation generation.
Deep networks can be designed to generate human-understandable explanations in a supervised manner.
Self-explanatory methods share several desirable aspects of xAI, but the authors particularly emphasize their inherent transparency.
By incorporating interpretable explanations at the core of its design, it avoids the common need for post-mortem explanation methods.
The resulting incomprehensible explanations may also provide natural insights into the plausibility of the predictions provided.
Self-explanatory deep learning has not yet been applied to chemical or drug design. Including interpretability by design can help bridge the gap between machine representation and human understanding of many types of problems in drug discovery.
xAI: Uncertainty Assessment
Uncertainty estimates, the quantification of errors in predictions, constitute another approach to model interpretation.
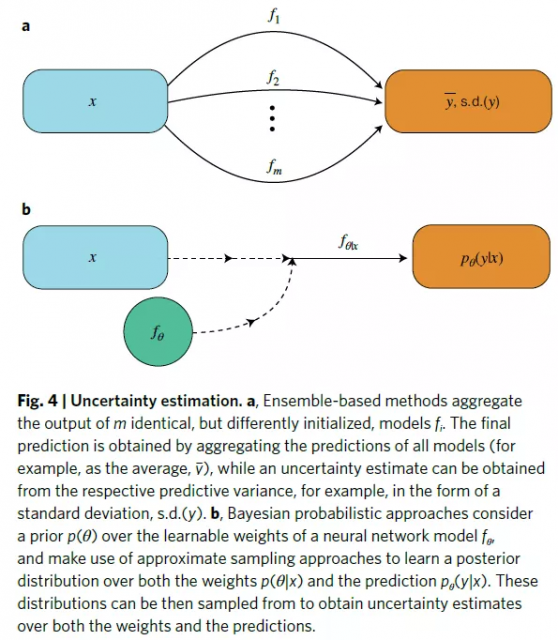
While some machine learning algorithms, such as Gaussian processes, provide built-in uncertainty estimates, deep neural networks are notoriously bad at quantifying uncertainty. This is one reason why there have been some efforts devoted to specifically quantifying uncertainty in neural network-based predictions. Uncertainty estimation methods can be grouped into the following categories.
inModel ensembles improve overall forecast quality and have become the standard for uncertainty estimation.
Probabilistic methods aim to estimate the posterior probability of a model output or to perform post-hoc calibration.
Other methods. The Lower Upper Limit Estimation (LUBE) method trains a neural network with two outputs corresponding to the upper and lower bounds of the prediction.
xAI: Available software
Given the current interest in deep learning applications, several software tools have been developed to facilitate model interpretation.
A prominent example is Captum, an extension to the PyTorch deep learning and automatic differentiation package that powers most of the feature attribution techniques described in this work.
Another popular package is Alibi, which provides instance-specific explanations for some models trained with scikit-learn or TensorFlow packages. Some of the explanation methods implemented include anchors, contrastive explanations, and counterfactual examples.
xAI:Conclusion and Outlook
In the context of drug discovery, full comprehensibility of deep learning models can be difficult to achieve, although the predictions provided can still prove useful to practitioners. In striving to find explanations that match human intuition, it will be critical to carefully design a set of controlled experiments to test machine-driven hypotheses and improve their reliability and objectivity.
Given the diversity of possible explanations and methods applicable to a task, current xAI also faces technical challenges. Most methods are not off-the-shelf, "out-of-the-box" solutions and require customization for each application.
Furthermore, a solid understanding of the domain problem is crucial to determine which model decisions require further explanation, which types of answers are meaningful to users, and which are trivial or expected.
For human decision-making, the explanations generated with xAI must be non-trivial, non-artificial, and sufficiently informative for the respective scientific community. Finding such a solution, at least for now, will require the combined efforts of deep learning experts, cheminformatics and data scientists, chemists, biologists and other domain experts to ensure that xAI methods work as intended and provide reliable answers.
Further exploration of the opportunities and limitations of existing chemical languages in representing the decision spaces of these models will be particularly important.
A step forward is to build on interpretable "low-level" molecular representations that have immediate implications for chemists and are amenable to machine learning.
Many recent studies rely on well-established molecular descriptors that capture a priori defined structural features. Often, molecular descriptors capture complex chemical information while being relevant to subsequent modeling.
Thus, when pursuing xAI, there is an understandable tendency to employ molecular characterizations that can be more easily rationalized in the language of known chemistry. The interpretability of the model depends on the chosen molecular representation and the chosen machine learning method.
With this in mind, the development of novel interpretable molecular representations for deep learning will constitute a key area of research in the coming years, including the development of self-explanatory methods to overcome impossibility by providing human-like explanations while providing sufficiently accurate predictions. Barriers to interpretive but informative descriptors.
Due to the current lack of methods encompassing all the desirable features of xAI outlined, consensus approaches will play a major role in the short and medium term, which combine the advantages of a single (X)AI approach and increase the reliability of the model. In the long run, the xAI approach will constitute an approach that provides multi-faceted vantage points for modeled biochemical processes by relying on different algorithms and molecular representations.
Currently, most deep learning models in drug discovery do not consider the applicability domain limit, the region of chemical space that satisfies statistical learning assumptions.
In the authors' opinion, these limitations should be considered as an integral part of xAI, since the evaluation of these limitations and the critical evaluation of model accuracy proved to be more beneficial to decision-making than the modeling method itself. Knowing when to apply which particular model will likely help address deep learning models' high confidence in mispredictions while avoiding unnecessary inferences.
Along these lines, in time- and cost-sensitive scenarios, such as drug discovery, deep learning practitioners have a responsibility to carefully examine and interpret the predictions that result from their modeling choices.
Given the current possibilities and limitations of xAI in drug discovery, it stands to reason that the continued development of hybrid approaches and alternative models that are easier to understand and more computationally powerful will not lose its importance.
Currently, xAI in drug discovery lacks an open community platform to share and improve software, model interpretations, and respective training data through the collaborative efforts of researchers from different scientific backgrounds. Initiatives like MELLODDY (Machine Learning Ledger Orchestration for Drug Discovery, melloddy.eu) for decentralized, federated model development and secure data processing among pharmaceutical companies are a first step in the right direction. This collaboration will hopefully facilitate the development, validation and acceptance of xAI and the associated explanations provided by these tools.


Comments