Development and Prospect of Integrated Deep Learning in Bioinformatics
In this issue, we will introduce the article "Ensemble deep learning in bioinformatics" published in Nature machine intelligence by Professor Jean Yang's research group at the University of Sydney.
This article reviews recent key developments in ensemble deep learning and how it can be applied to the field of bioinformatics.
At the same time, the authors also detail the research, development, and challenges of ensemble deep learning from basic sequence analysis to systems biology.
1. Deep learning: Main idea
Ensemble and deep learning have been regarded as two separate approaches in the field of bioinformatics.However, these two technologies have developed rapidly in recent years.
Many researchers have found that the integrated deep learning model has superior performance when dealing with small samples, high dimensions, and unbalanced distribution of data.
Therefore, more and more people begin to use the Eyes turn to the field of ensemble deep learning.
In the field of bioinformatics, both ensemble learning and deep learning methods have been widely studied and reviewed, but the application of ensemble deep learning in the biomedical field has not been documented yet.
This article reviews the fundamentals of ensembles and deep learning, and summarizes and categorizes recent developments in ensemble deep learning. In addition, the authors survey the application of integrated deep learning in bioinformatics, and then discuss the challenges and opportunities in this area to facilitate future research and development across multiple disciplines.
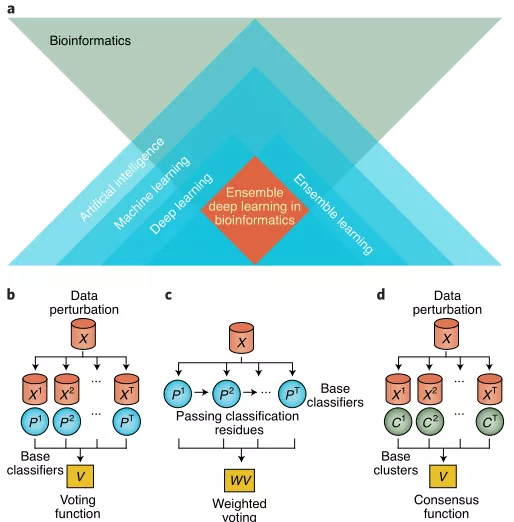
Figure 1 shows the focus of this paper and some classic ensemble learning methods.
2.Deep learning: Related research
2.1 Fundamentals of Ensemble and Deep Learning
Ensemble learning is the combination of multiple "base" models to perform tasks such as supervised and unsupervised learning.Classic supervised learning ensemble methods fall into three categories: bagging-based methods, boosting-based methods, and stack-based methods. Traditional unsupervised ensemble learning also relies on the ensemble of base models. The principle of the ensemble approach is "many is better than one".
The most basic architecture of deep learning is a densely connected neural network (DNN), which consists of a series of neurons, each layer is connected to all neurons in the previous layer.
Models like CNN, RNN, ResNet, etc. are all developed on the basic architecture.
2.2 Integrated Deep Learning
Deep learning often has high variance and may get stuck in local loss minima during training, and the method of integrating multiple deep learning models has better generalization ability than a single model.The article categorizes and summarizes supervised and unsupervised ensemble deep learning strategies.
Supervised ensemble deep learning can be broadly classified into three categories: ensembles across multiple models, ensembles of single models, and ensembles of model branches.
Ensembling across multiple models is usually the direct aggregation of multiple independent models to facilitate the diversity of the underlying network.
Complementary learning on training data can achieve better ensemble generalization, or multiple-choice learning can specialize on a specific subset of data.
In "implicit ensemble", a single neural network can achieve an effect similar to integrating multiple networks. When training a single neural network, a technique is used to randomly activate layers of neurons so that networks with different architectures are implicitly integrated.
As in ResNets, the ResBlocks building blocks are randomly deactivated. Compared with multiple model ensembles, a single model ensemble reduces training costs, but at the same time may reduce model diversity.
Therefore, the model branch ensemble is to share the lower layer and add the branch layer. By sharing information, it avoids searching for parameters from scratch, and the convergence speed is faster.
Most unsupervised ensemble deep learning methods employ autoencoders. Similar to supervised methods, unsupervised ensemble methods can be divided into methods that generate and combine multiple models through data and model perturbations, and methods that achieve implicit ensemble within a single model.
Typical ensemble deep learning frameworks in supervised and unsupervised learning are sequentially shown in Figure 2:
3.Deep learning: Application of ensemble deep learning in biomedical field
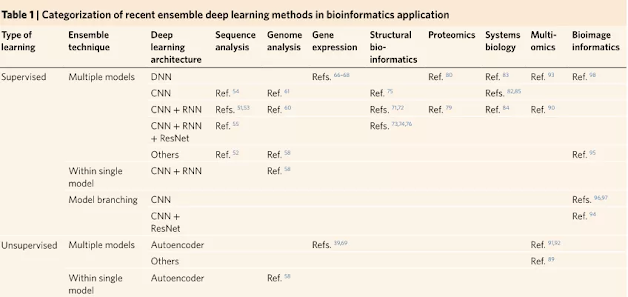
The article categorizes representative work in different fields of bioinformatics applications and identifies their advantages, such as improved model accuracy, reproducibility, interpretability, and model inference.The summary results of the article are shown in Table 1:
4.Deep learning: Challenges and opportunities
Ensemble deep learning is significantly better than deep learning in terms of small samples, high dimensionality and hierarchy imbalance, data noise and heterogeneity, model interpretability, network architecture selection, and computational cost. In the field of biomedicine, there are usually a series of problems such as small sample size and high data dimension. Therefore, it is a good development direction to use deep ensemble learning to solve problems in the field of biomedicine.
The development of ensemble deep learning has greatly enriched the field of deep learning with novel architectures and ensemble strategies, improving the accuracy, reliability, and efficiency of models, and robustness to small samples, high dimensions, and data noise in bioinformatics applications. Significant and widespread breakthroughs have been made in different fields.
Today, the development and application of models capable of explaining biological systems is still in its infancy, and there is still much room for research in ensemble deep learning.



Comments